Obtaining transconjugant strains for bacteria-rice interaction studies
Main Article Content
Abstract
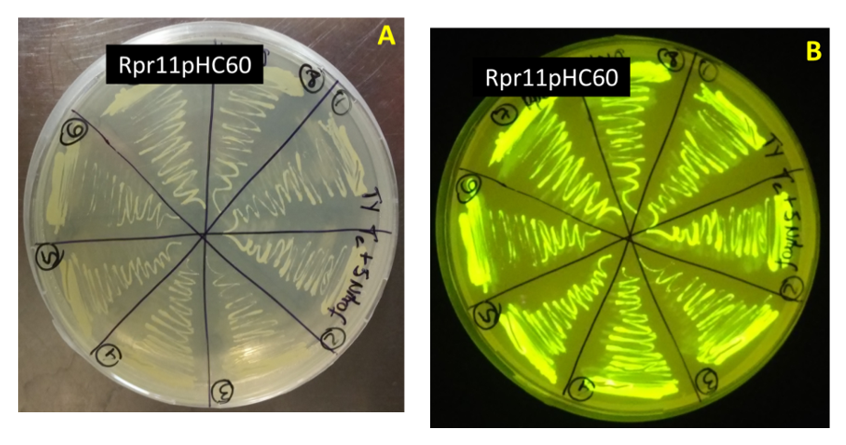
The objective of this work was to obtain transconjugant strains for the study of plant-bacteria interaction. The obtaining of transconjugant bacteria with the plasmid pHC60 was carry out by triparental mating with two Escherichia coli strains and two strains isolated from Cuban rice cultivars. Furthermore, an inoculation assay with rice plants cv. INCA LP-5 and transconjugant bacteria was carry out and confocal fluorescence microscopy were used to locate the bacteria in plant tissues. Two transconjugant strains containing gfp gene were obtained, since they produced fluorescent colonies in ultraviolet light, in culture medium with tetracycline and 5-nitrofurantoin. The multiplication dynamics of wild type and transconjugant strains don’t showed differences in the end of bacteria growth in the medium. Two transconjugant strains were showed colonizing rice root, one of them as a putative endophyte.
Article Details

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Those authors who have publications with this journal accept the following terms of the License Attribution-NonCommercial 4.0 International (CC BY-NC 4.0):
You are free to:
- Share — copy and redistribute the material in any medium or format
- Adapt — remix, transform, and build upon the material
The licensor cannot revoke these freedoms as long as you follow the license terms.
Under the following terms:
- Attribution — You must give appropriate credit, provide a link to the license, and indicate if changes were made. You may do so in any reasonable manner, but not in any way that suggests the licensor endorses you or your use.
- NonCommercial — You may not use the material for commercial purposes.
- No additional restrictions — You may not apply legal terms or technological measures that legally restrict others from doing anything the license permits.
The journal is not responsible for the opinions and concepts expressed in the works, they are the sole responsibility of the authors. The Editor, with the assistance of the Editorial Committee, reserves the right to suggest or request advisable or necessary modifications. They are accepted to publish original scientific papers, research results of interest that have not been published or sent to another journal for the same purpose.
The mention of trademarks of equipment, instruments or specific materials is for identification purposes, and there is no promotional commitment in relation to them, neither by the authors nor by the publisher.
References
Sarhan MS, Hamza MA, Youssef HH, Patz S, Becker M, ElSawey H, et al. Culturomics of the plant prokaryotic microbiome and the dawn of plant-based culture media–A review. Journal of Advanced Research. 2019;19(19):15-27. doi: 10.1016/j.jare.2019.04.002
Hartmann A, Fischer D, Kinzel L, Chowdhury SP, Hofmann A, Baldani JI, et al. Assessment of the structural and functional diversities of plant microbiota: Achievements and challenges–A review. Journal of Advanced Research. 2019;19:3-13. doi: 10.1016/j.jare.2019.04.007
Geddes, BA, Paramasivan P, Joffrin A, Thompson AL, Christensen K, Jorrin B, et al. Engineering transkingdom signalling in plants to control gene expression in rhizosphere bacteria. Nature communications. 2019;10(1):1-11. doi: 10.1038/s41467-019-10882-x
Chen X, Miché L, Sachs S, Wang Q, Buschart A, Yang HY, et al. Rice responds to endophytic colonization which is independent of the common symbiotic signaling pathway. New Phytologist. 2015;208(2):531-543. doi: 10.1111/nph.13458
Wu Q, Peng X, Yang M, Zhang W, Dazzo FB, Uphoff N, et al. Rhizobia promote the growth of rice shoots by targeting cell signaling, division and expansion. Plant Molecular Biology. 2018;97(6):507-523. doi: 10.1007/s11103-018-0756-3
Hernández I, Taulé C, Pérez-Pérez R, Battistoni F, Fabiano E, Rivero D, et al. Endophytic rhizobia promote the growth of Cuban rice cultivar. Symbiosis. 2021a; 85(2):1-16.
doi: 10.1007/s13199-021-00803-2
Hernández I, Pérez-Pérez R, Nápoles MC, Maqueira LA, Rojan O. Rhizospheric rhizobia with potential as biofertilizers from Cuban rice cultivars. Agronomía Colombiana. 2021b; 39(1),26-37. doi: 10.15446/agron.colomb.v39n1.88907
Hanahan D. Studies on transformation of Escherichia coli with plasmids. Journal of Molecular Biology. 1983;166(4):557-580. doi: 10.1016/s0022-2836(83)80284-8
Ditta G, Stanfield S, Corbin D, Helinsky D. Broad host range DNA cloning system for gram-negative bacteria: construction of a gane bank of Rhizobium meliloti. Proceedings of National Academy of Sciences. 1980;77(12):7347-7351. doi: 10.1073/pnas.77.12.7347
Padda KP, Puri A, Zeng Q, Chanway CP, Wu X. Effect of GFP-tagging on nitrogen fixation and plant growth promotion of an endophytic diazotrophic strain of Paenibacillus polymyxa. Botany. 2017; 95(9):933-942. doi: 10.1139/cjb-2017-0056
Jiménez-Gómez A, Flores-Félix JD, García-Fraile P, Mateos PF, Menéndez E, Velázquez E, et al. Probiotic activities of Rhizobium laguerreae on growth and quality of spinach. Scientific Reports. 2018;8(1):1-10. doi: 10.1038/s41598-017-18632-z
Ferreira NS, Matos GF, Meneses CH, Reis VM, Rouws JR, Schwab S, et al. Interaction of phytohormone-producing rhizobia with sugarcane mini-setts and their effect on plant development. Plant and Soil. 2020; 451(1):221-238. doi: 10.1007/s11104-019-04388-0